Main Menu
Welcome
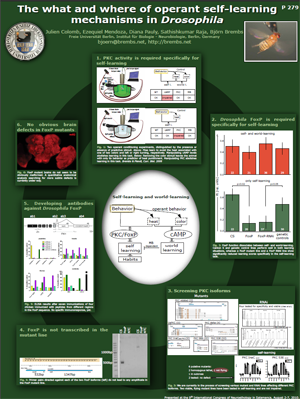
The fruit fly Drosophila is also capable of operant learning, albeit not of speech or song. Instead, we can get them to compare the consequences of their flight maneuvers with a desired outcome. For instance, we can make turning off of a punishing heat-beam the desired outcome and couple it to certain turning attempts. To do this, we tether the flies and measure their turning behavior using a torque meter:
Because the procedure is conceptually analogous to speech and song learning, we hypothesized that if there is an orthologue of FOXP2 in flies, it should be involved in this form of operant learning. Last year (or was it already two years ago?) I was expressing this hypothesis over beers at a conference to my good friend Troy Zars, who told me he had recently gotten hold of a mutant line of what he thought was FoxP in flies. When I received the flies back home, I tested them and the couple of mutants I tested the first day they learned just fine and I thought the hypothesis wasn't as great as I had initially hoped. Nevertheless, a day is nothing so I tested the flies for a few more days and lo and behold, the average learning score was not significant, whereas the control line showed nice learning. What was even more convincing was that other forms of learning, which did not require the animals to learn about their behavior, were not impaired in the mutants. This was analogous to the results we had when manipulating PKC and which has led us to term the two learning mechanisms self-learning and world-learning, respectively.
Not content with a single experiment, we decided to use a different method besides mutation to get at the gene, RNAi. We used the genetic toolbox of Drosophila and knocked down FoxP. We did this in two separate replicates where the driver and effector genes came from father or mother, respectively. Both crosses showed the same result as the mutant flies: world-learning was not affected, but self-learning was impaired. With three independent replicates and two different methods, we now feel fairly confident that FoxP is required for self-learning in flies. Moreover, the evolutionary relationship to the vertebrate FOXP2 makes for a plausible explanation of why this gene should be involved in this kind of learning.
The genetics of FoxP in Drosophila were by no means simple, though. In fact, the mutant line was not in what the databases told us was FoxP, but in a small, neighboring 'gene'. Further, the RNAi line we used was directed towards this 'gene', as this is where the mutation was. You can see the arrangement in FlyBase: FoxP is CG16899 and the other 'gene' is CG32937. There was a lot of bioinformatics data suggesting that the small 'gene' was in fact a part of FoxP and that the database was simply not correct, but for a publication we would have to show that experimentally as well. So we initiated some small side projects to study the expression of these genes on the mRNA and the protein level: PCR to look for transcripts and antibodies to localize the protein.
Two days ago, just after the PCR and cloning experiments on the transcripts of the two genes were starting to take off and we were getting the first results, we were alerted to a publication that had just been published, which was confirming our suspicions: CG32937 was indeed part of FoxP! From their paper:
Our in silico analysis suggested that CG16899 could include an extra exon at its 3´ extremity, to date reported as a separate gene (CG32937). This prediction stems from the nature of the predicted protein encoded by CG32937 that showed forkhead domain like features. The alignment of the predicted protein from CG32937 to exon 7 of CG16899, revealed a 60% amino acid identity. These results suggest that the predicted locus CG32937 could be instead an additional exon of CG16899 that would be alternatively spliced, joining with exon 6 to form two different, yet recognizable, forkhead domains
So, all of a sudden, we were cast many months forward in our project. Luckily, we hadn't performed many of these experiments, yet, but our plan had been very similar to what Santos et al. have just published.However, now everybody knows that CG16899 is FoxP and given the popularity of FOXP2 (I don't think there are too many genes with their own Wikipedia page), other researchers may have the same hypothesis I had last year. This means we now have to move quickly and try and finish up and publish our results. For now, this blog post will claim precedence and we will present the data on a poster next week at the International Congress of Neuroethology in Salamanca, Spain, along with our other, unrelated poster. In November, we will present these results at the Society for Neuroscience conference, but I hope we have a first draft of a manuscript about ready by then.
My interpretation of these results is that the transcription factor FoxP had evolved in the Urbilaterian, the last common ancestor of humans and flies, as a gene important for self-learning. In the vertebrate lineage, the gene underwent several duplications and specializations until FOXP2 became involved in speech/language.
The exciting thing is that besides PKC and FoxP, virtually nothing is known about the mechanisms of self-learning. It is not known what form of neuronal plasticity it is, where in the brain it is implemented or why it seems so exquisitely separated from other forms of learning. Plenty of low hanging fruit in this field! Let's get to work!

Santos, M., Athanasiadis, A., Leitao, A., DuPasquier, L., & Sucena, E. (2010). Alternative splicing and gene duplication in the evolution of the FoxP gene sub-family Molecular Biology and Evolution DOI: 10.1093/molbev/msq182
Fisher, S., & Scharff, C. (2009). FOXP2 as a molecular window into speech and language Trends in Genetics, 25 (4), 166-177 DOI: 10.1016/j.tig.2009.03.002
Posted on Friday 30 July 2010 - 19:02:06 comment: 0
{TAGS}
{TAGS}
You must be logged in to make comments on this site - please log in, or if you are not registered click here to signup
Render time: 0.0946 sec, 0.0057 of that for queries.